As more sophisticated algorithmic approaches demonstrate greater accuracy, diverse datasets become more accessible and technical computing power grows, the use of machine learning (ML) techniques in drug discovery is shifting from theoretical possibility to real world utility. One example is the recent success of MIT researchers who used ML to discover a new class of compounds effective in killing antibiotic-resistant bacteria. As structural diversity is limited in antibiotic innovation, given the small number of mechanisms these drugs can target, the ability of ML to identify unexpected drug-like candidates with activity was a huge step forward.
Though exciting, that outcome is still rare. However, more commonly ML is enabling researchers to screen large sets of potential therapeutic compounds to identify those predicted to be most potent relevant to targets of interest. This in silico prioritization of candidates for synthesis and testing significantly reduces the cost per lead for drug discovery teams by shrinking the pool of molecules that are prepared or purchased.
These benefits, and even greater aspirations for ML in drug discovery, can only be realized on a larger scale, however, if algorithms can be relied upon to consistently deliver accurate predictions of bioactivity. What can we augment the ML approach with to increase the accuracy and reliability of those predications? As demonstrated by research using CAS substance data recently published in the Journal of Chemical Information and Modeling, the answer might surprise you; it’s a human chemist.
Read the full journal article Impact of Chemist-In-The-Loop Molecular Representations on Machine Learning Outcomes to see the data showing how chemist-curated molecular fingerprints impacted prediction accuracy.
Data, descriptors and algorithms: The trifecta that drives predictive success
Algorithms are often thought of as the most important component of ML, and obviously, they are critical. Extensive energy is dedicated to building, testing and optimizing algorithmic approaches to model each situation of interest. However, I would argue that data quality remains the single most import factor for building reliable ML models. When, due to limitations of availability, curation or diversity, the available data cannot accurately reflect the universe of possibilities that the algorithm should consider, the sophistication of the algorithm is wasted. To fuel an algorithmic approach to assessing potential drug candidates, it is very important to have a comprehensive, clean set of structural, biological and physical properties. The CAS REGISTRY®, which currently contains data on more than 166 million small molecules curated by scientists from over 100 years of published scientific literature and patents, serves as an excellent data foundation for this type of work.
There is, however, one more critical component to predictive chemistry that has been long overlooked, but more recently is getting greater attention, molecular descriptors. Also commonly known as molecular fingerprints, as their name suggests molecular descriptors describe key features of each chemical molecule to the algorithm. Some of the thousands of potential features of a candidate molecule include number of atoms, atom type and bond configuration. However, the features that are most relevant to predictive outcomes vary depending on the goal of the algorithm. Despite this, most ML efforts today rely on a generic set of molecular descriptors. Some of the most popular descriptors (included in the Extended Connectivity Fingerprints) are based on the Morgan algorithm, developed at CAS in the early 1960s by Harry Morgan. Though these are a good starting point, our research has demonstrated that an enhanced fingerprint recently developed by CAS that includes over 25,000 structural features selected by our team of chemists consistently improves the accuracy of bioactivity predictions.
Turning chemists into tailors produces better fitting predictions
Our recently published research focused on comparing the accuracy of bioactivity predictions using a number of common generic descriptors with those using these new descriptors developed by leveraging the expertise of CAS chemists to add additional feature richness suitable for many ML applications. For simplicity, we referred these chemist-curated descriptors as CAS fingerprints. The study results show that CAS fingerprints, when used for predicting bioactivity for a large benchmark set of 88 diverse targets, outperform commonly used molecular descriptors such as the ECFP (Morgan), Avalon, Atom Pair and Topological Torsion fingerprints. Based on ROC-AUC and PRC-AUC, the proprietary CAS fingerprint had the highest average rank in random forest ML models.
Preliminary testing shows that additional accuracy gains can be achieved when chemists further customize the features used in these broadly applicable CAS-enhanced fingerprints for each individual algorithmic application. These customized fingerprints are created by selecting the most informative features for the targets of interest. Various dimensionality reduction techniques such as principle component analysis can be used to further improve accuracy, stability and scalability of the predictive models. Feature importance analysis can also be used to gain additional insights into elements most relevant to biological activity, creating a virtuous loop of optimization.
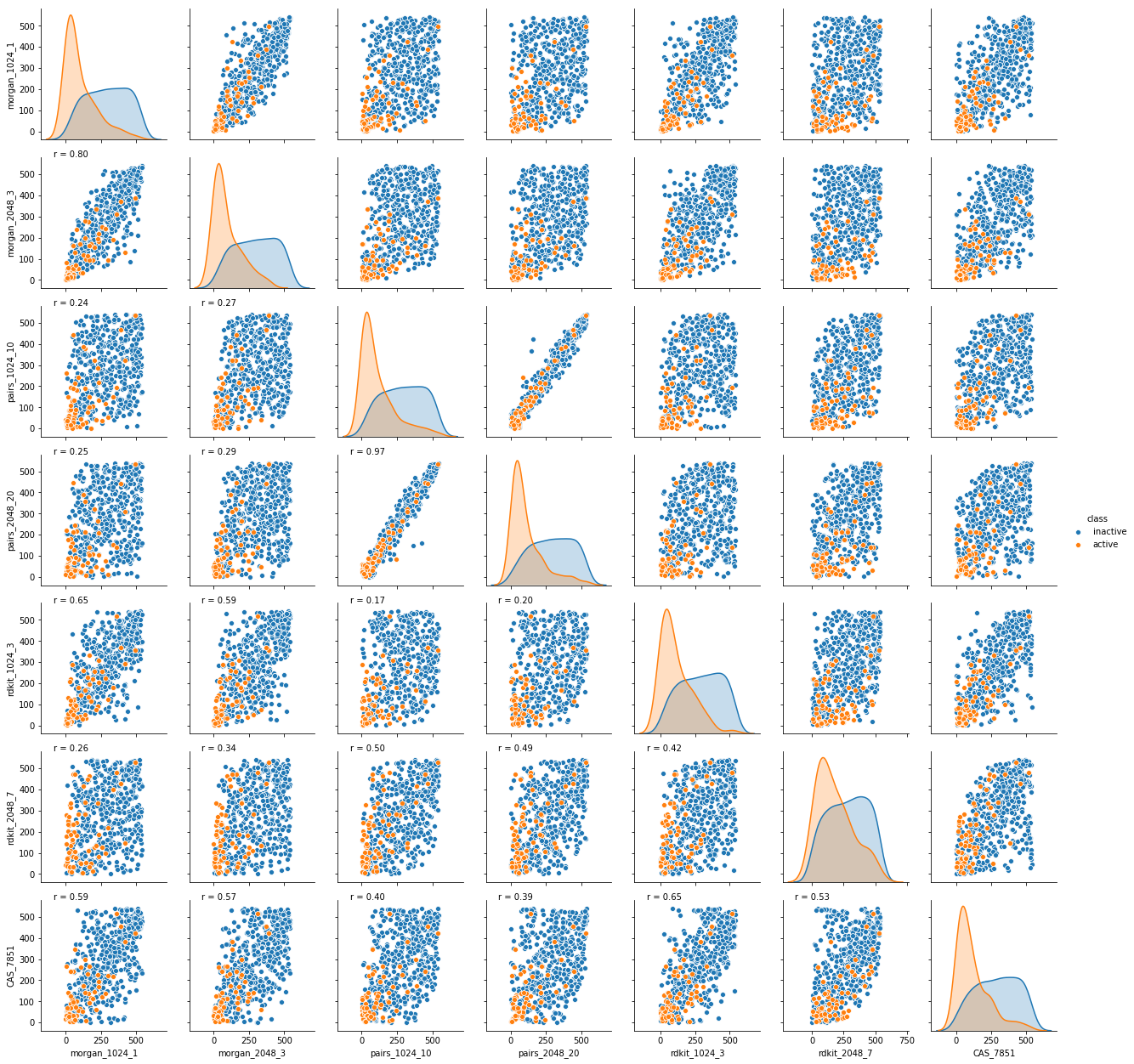
Though these initial accuracy gains and further possibilities are exciting, potentially the most interesting finding of this work is the demonstrated impact of the CAS fingerprint on the diversity of predictive outcomes. This highlights their potential to also positively impact innovation. The CAS fingerprint frequently finds active structures that are drastically different from those predicted by models built with more generic, common molecular descriptors. As can be seen from Figure 1 below, the correlation between CAS fingerprint and other tested models is very low. Therefore, the CAS fingerprint captures orthogonal chemical signals that provide unique insights not provided by other commonly used molecular descriptors.
Broader applications of enhanced molecular descriptors
Customized molecular descriptors have additional applications as part of a scaled-up, ML-enabled R&D workflow. For example, in the early stages discovery, it is highly desirable to identify a structurally diverse set of compounds that have similar activity but contain different core structures (i.e., scaffold hopping), as structurally novel drugs have been more than twice as likely to be granted breakthrough therapy designation status by the FDA. Scaffold-hopping potential is considered an important ability for ML methods. However, the potential to retrieve structurally diverse molecules varies by fingerprint. Preliminary analysis has shown the CAS fingerprint has better scaffold-hopping potential than other commonly used fingerprints. This is an important factor for discovering entirely new classes of candidates or accurately assessing the activity of structurally diverse candidate pools.
The ML-supported screening approach described above also can be used to proactively screen all new compounds entering internal as well as external data sets, including CAS REGISTRY, for potential activity against an organization’s portfolio of priority targets. By organizing these target-specific ML models into pipelines, this approach can continuously feed the most potent candidates into the pipeline. Such use cases are also not limited to drug delivery. Approaches discussed here that rely on ML for identifying, screening and prioritizing candidate compounds are also being adopted in other chemical applications such as the development of novel pesticides.
Do you have thoughts on other impactful applications of enhanced or customized molecular descriptors in drug discovery or other chemical applications? Share your thoughts in the comments below or connect with our Custom Services team.

