Gain new perspectives for faster progress directly to your inbox.

RAS proteins: an elusive target?
Around one in every five human cancers have at least one form of RAS mutation (K-RAS, H-RAS, and N-RAS), making RAS the most frequently mutated gene family in human cancers. RAS proteins, located on the cell plasma membrane, act as molecular switches sending signals for cell growth. However, mutations in RAS proteins may cause them to be constantly active and send growth signals uncontrollably, which leads to abnormal cell proliferation and cancer formation.
Despite their prolificacy, there is a distinct lack of therapies that target these RAS proteins. RAS inhibitors have been investigated in cancer treatment for more than three decades, yet RAS proteins have come to be known as “undruggable” due to their elusive inhibitory potential – that is, until recently. Earlier this year the FDA approved sotorasib (developed by Amgen and commercially known as Lumakras™) for the treatment of lung cancer – the first RAS inhibitor to be approved as a therapy.
The approval of sotorasib is a significant step forward in RAS inhibition, and research and development efforts in discovering further RAS inhibitors has intensified. Here, we explore how the gap is closing on RAS proteins – what were once considered elusive, undruggable targets are now emerging as a promising cancer treatment.
K-RAS: the most common mutation in RAS genes
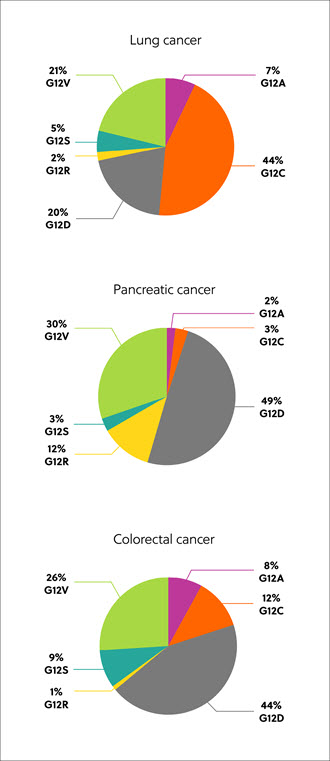
The RAS genes code for proteins, which exist in four isoforms: K-RAS4A, K-RAS4B, N-RAS, and H-RAS. Mutated RAS isoforms, codon, and amino acid substitution vary by tissue and cancer type, but the most common are mutations in the K-RAS isoform – found in approximately 22% of RAS-mutated cancers. Eighty percent (80%) of K-RAS mutations occur at amino acid position 12, from glycine to other residues, including cysteine (G12C, 14%), aspartic acid, (G12D, 36%), and valine (G12V, 23%) (Figure 1).1
Discovery of RAS proteins as candidates for cancer treatment
RAS inhibitors can be identified by using X-ray crystallography. Using this method, RAS protein structures can be examined to identify potential binding pockets for small molecules to occupy within human cancer cells. This type of approach — structure-based drug design — makes it possible to discover hundreds of chemical substances that can potentially bind within specific pockets. The potential RAS inhibitors usually consist of a scaffold structure that is slightly modified with a variety of functional groups to enhance activity, selectivity, and decrease toxicity. This results in lead compounds that can be further analyzed, enhanced, and tested with the hope of evaluating them in human cancer clinical trials.
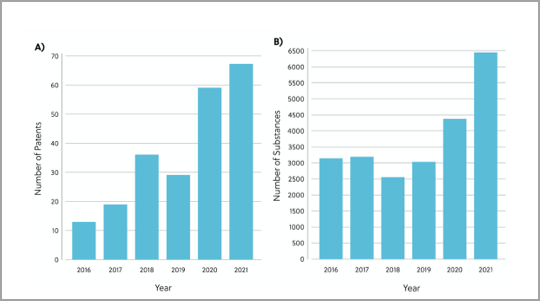
To gain a deeper understanding of the current RAS inhibitor landscape, we’ve reviewed the patents and publications related to RAS inhibitors in the CAS Content Collection™. The analysis revealed 26,958 chemical substances with therapeutic or pharmacological roles in the direct RAS inhibitory space. The number of chemical substances and patents in this space have increased per year, underpinning how research interest and efforts towards RAS inhibitor discovery are gaining pace (Figure 2).
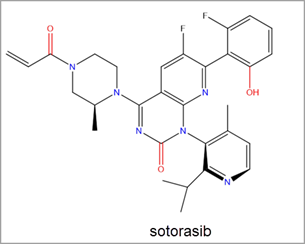
The recent FDA approval of Amgen’s sotorasib led to a significant uptick in research efforts towards RAS inhibitor discovery. Sotorasib is a covalent inhibitor of KRAS G12C via the Switch-II pocket. It was the first KRAS inhibitor to be approved for use in human cancer treatment and to treat KRAS G12C mutated non-small cell lung cancer (NSCLC) (Figure 3).2
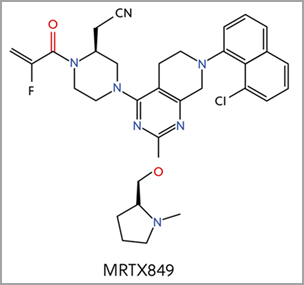
Four additional KRAS-G12C inhibitors are currently in clinical trials, including MRTX849, which is based on a similar core structure to sotorasib (Figure 4)2. Different functional groups resulted in different binding mechanisms to key elements of the Switch II pocket. MRTX849 earned FDA breakthrough therapy designation for KRAS G12C positive NSCLC in June 2021.
The journey continues: expanding the breadth of direct RAS inhibitor targets
As more molecules binding RAS are discovered, more surfaces on RAS isoforms and RAS proteins are being identified as potential small molecule targets.
As mutated RAS isoforms, codon, and amino acid substitution vary by tissue and cancer type, varying approaches are needed from the current G12C inhibitors to increase the range of cancer therapies. Future opportunities include expanding the type of amino acids that can be targeted by inhibitors, such as G12D and G12V, which may broaden the types of cancer we are able to treat.
The gateway is now open for RAS inhibition and by gaining a better understanding of RAS oncoprotein structure and binding pocket configuration for small-molecule targets, novel RAS inhibitors can be developed and enhanced for optimal activity in RAS mutated cancers.
Read our white paper to find out more about the continuing journey in uncovering RAS targets, including a more detailed overview of the current RAS inhibitor landscape of chemical structures and future opportunities.
References
1. H. Chen et al., Small-molecule inhibitors directly targeting KRAS as anticancer therapeutics. J. Med. Chem. 63 (2020) 11404–14424. doi: 10.1021/acs.jmedchem.0c01312.
2. L. Goebel et al., KRASG12C inhibitors in clinical trials: a short historical perspective. RSC. Med. Chem. 11 (2020) 760. doi: 10.1039/d0md00096e.