Gain new perspectives for faster progress directly to your inbox.
For years, biologists believed the amino acid sequence of each protein determines its three-dimensional structure, which, in turn, determines its function. However, there is a large group of proteins and regions that lack a fixed or ordered 3D structure, yet they still exhibit essential biological activities—so-called intrinsically disordered proteins.
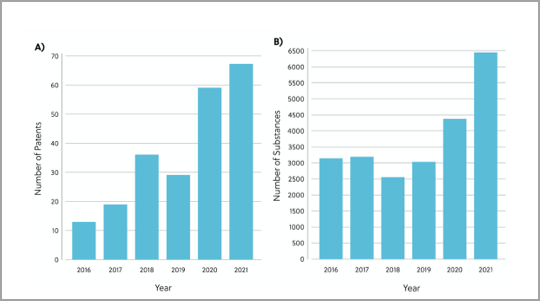
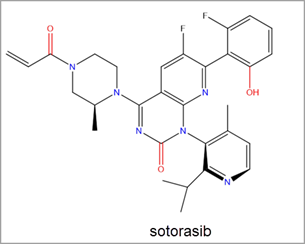
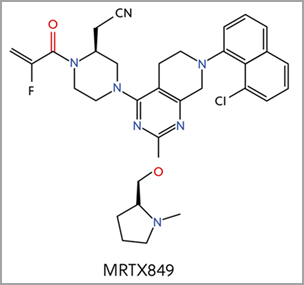
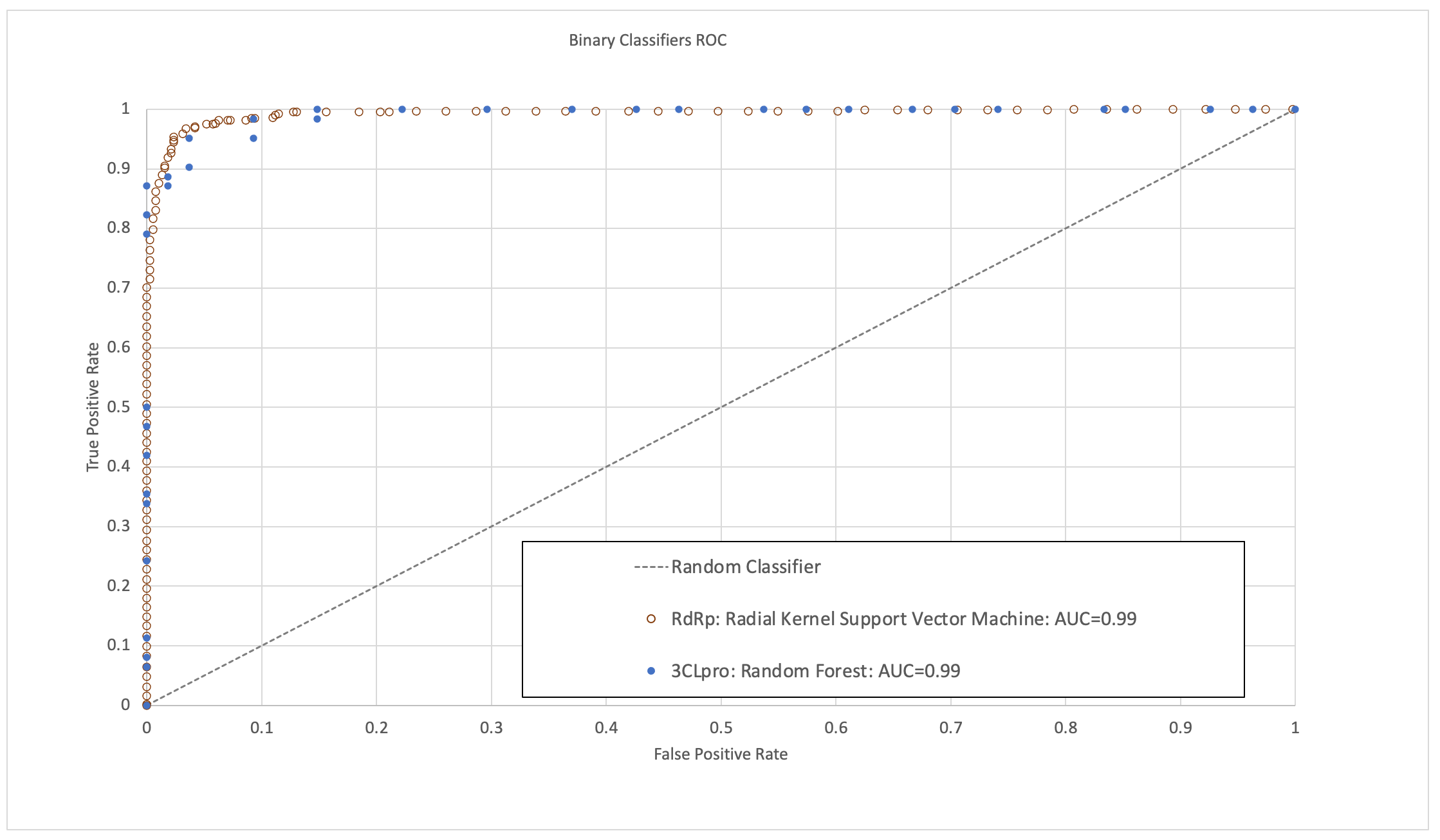
It turns out that these intrinsically disordered proteins may be the key to better overcoming diseases in neurodegeneration, diabetes, cardiovascular disease, amyloidosis, genetic diseases, and cancer. This peer-reviewed journal published in ACS Infectious Diseases reveals a landscape analysis of this emerging topic and identifies critical insights across therapeutic areas, from SARS-CoV2 to genetic diseases and cancers. The deep dive within discoveries around intrinsically disordered proteins and the opportunities ahead will enable faster progress for future therapies. Read the full publication here.